Transcript elongation by RNA polymerase II (RNAPII), once regarded as the simple extension of the initiated mRNA, is a complex and highly regulated phase of the transcription cycle. Many factors have been identified that contribute to the dynamic control of the elongation stage of transcription. There are transcript elongation factors (TEFs) that modulate the activity of RNAPII (green symbols in the scheme), while others facilitate the transcription through chromatin (blue and orange symbols), or covalently modify nucleosomal histones (i.e. methylation, acetylation, ubiquitination) within the transcribed regions (yellow symbols) (Van Lijsebettens and Grasser, 2014 (externer Link, öffnet neues Fenster)). Several TEFs including TFIIS, FACT, SPT4/SPT5 and PAF1C were found to associate with the elongating RNAPII (S2P, S5P phosphorylated) forming the transcript elongation complex (Antosz et al., 2017 (externer Link, öffnet neues Fenster)).
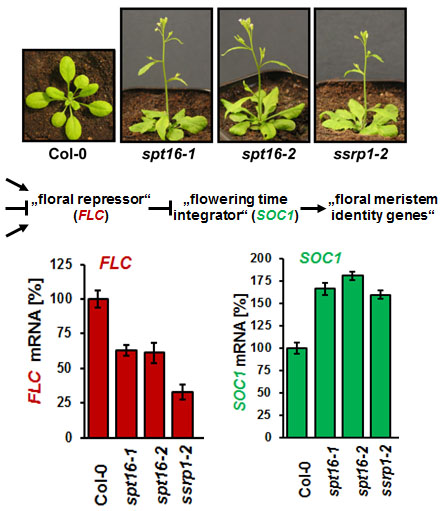
The Arabidopsis FACT (facilitates chromatin transcription) complex is composed of the SSRP1 and the SPT16 proteins and localises to the transcriptionally active euchromatin of the majority of cell types. The FACT histone chaperone is found to associate with the transcribed region of active genes in a transcription-dependent manner, suggesting that the FACT complex acts as transcript elongation factor assisting transcription in the chromatin context (Grasser, 2020 (externer Link, öffnet neues Fenster)). Arabidopsis mutant plants (ssrp1, spt16) that express reduced amounts of FACT subunits display various developmental defects including elevated number of smaller leaves, early bolting/flowering, and a bushy appearance due to reduced apical dominance. In addition, the architecture of mutant flowers is affected resulting in severely reduced seed production (Lolas et al., 2010 (externer Link, öffnet neues Fenster)). The early bolting of the mutant plants is associated with reduced transcript levels of the floral repressor FLC and consistently increased expression of SOC1.
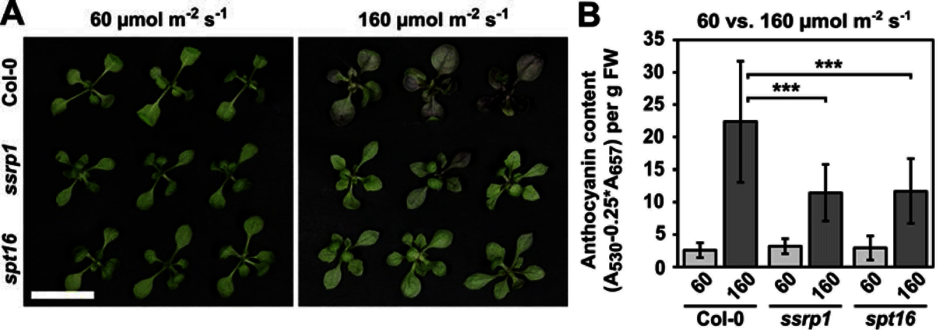
Seed germination analyses revealed that FACT contributes to the regulation of seed dormancy by promoting the expression of the DOG1 gene (Michl-Holzinger et al., 2019 (externer Link, öffnet neues Fenster)). Thus, FACT facilitates expression of DOG1 and FLC adjusting the crucial developmental transitions from seed dormancy to germination and from vegetative to reproductive development, respectively. Another study demonstrated that upon exposure to moderate high-light stress several genes encoding enzymes of the anthocyanin biosynthetic pathway are up-regulated to a lesser extent in ssrp1/spt16 compared to wild type plants. Accordingly, the mutant plants accumulate lower amounts of the purple anthocyanin pigments (Pfab et al., 2018 (externer Link, öffnet neues Fenster)). These findings indicate that FACT is a novel factor required for the accumulation of anthocyanins when the plants are exposed to environmental stress. The acidic region of the FACT subunit SPT16 is phosphorylated by protein kinase CK2 and the phosphorylation is required for establishing the correct nucleosome occupancy at transcriptional start sites of a subset of genes (Michl-Holzinger et al., 2022 (externer Link, öffnet neues Fenster)).
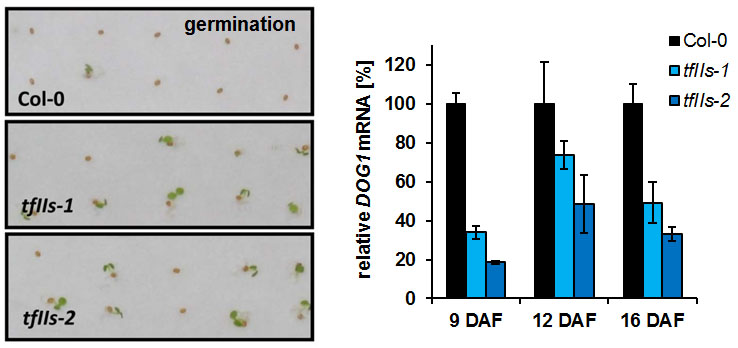
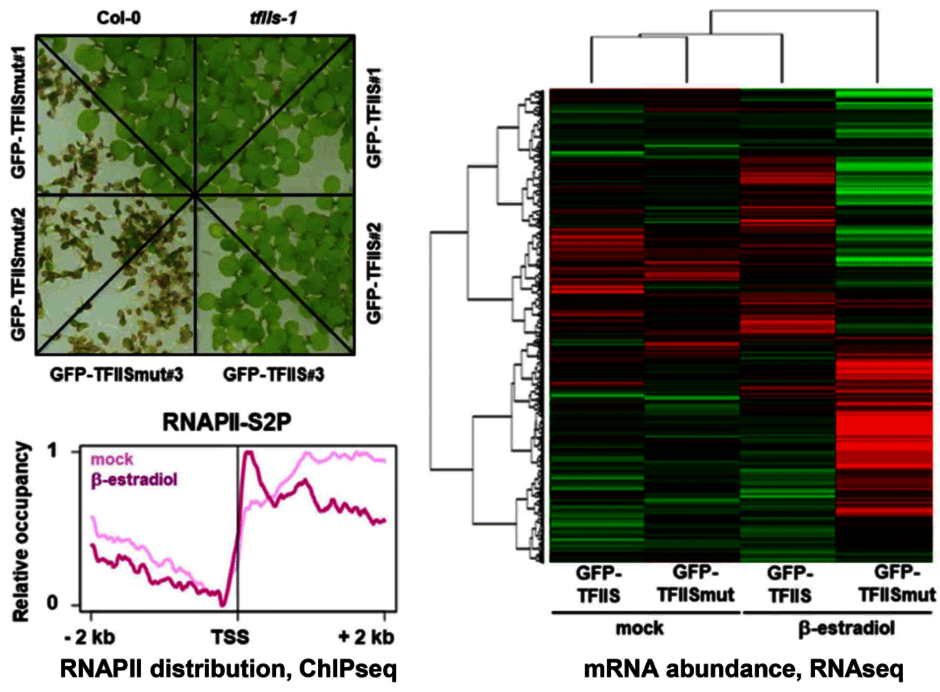
Another TEF that we are studying is TFIIS, an RNAPII-associated factor that can facilitate RNAPII transcription through sites that cause transient pausing or permanent arrest of transcribing RNAPII. Arabidopsis TFIIS can partially complement the phenotype of yeast cells lacking the endogenous TFIIS, indicating that Arabidopsis TFIIS plays a role in transcript elongation comparable to its yeast/mammalian counterparts. Mutant Arabidopsis plants develop normal, but the mutant seeds are severely affected in seed dormancy (Grasser et al., 2009 (externer Link, öffnet neues Fenster)). Thus, tfIIs seeds of fully elongated but still green siliques germinate efficiently, whereas comparable Col-0 seeds hardly germinate. The dormancy defect of tfIIs seeds is caused by reduced expression of the DOG1 gene encoding a potent regulator of seed dormancy (Mortensen and Grasser, 2014 (externer Link, öffnet neues Fenster)).
The elongation factor TFIIS stimulates the weak intrinsic transcript cleavage activity of RNAPII, which is required for efficient rescue of backtracked/arrested polymerase. A TFIIS mutant variant (TFIISmut) lacks the stimulatory activity to promote RNA cleavage, but instead efficiently inhibits unstimulated transcript cleavage by RNAPII. Constitutive expression of TFIISmut is lethal in Arabidopsis tfIIs plants. Induced, transient expression of TFIISmut in tfIIs plants provoked severe growth defects, transcriptomic changes and massive, transcription-related redistribution of elongating RNAPII within transcribed regions towards the transcriptional start site. The predominant site of RNAPII accumulation overlapped with the +1 nucleosome, suggesting that upon inhibition of RNA cleavage activity RNAPII arrest prevalently occurs at this position (Antosz et al., 2020 (externer Link, öffnet neues Fenster)). These findings suggest that polymerase backtracking/arrest frequently occurs in plant cells and RNAPII-reactivation is essential for correct transcriptional output and proper growth/development.
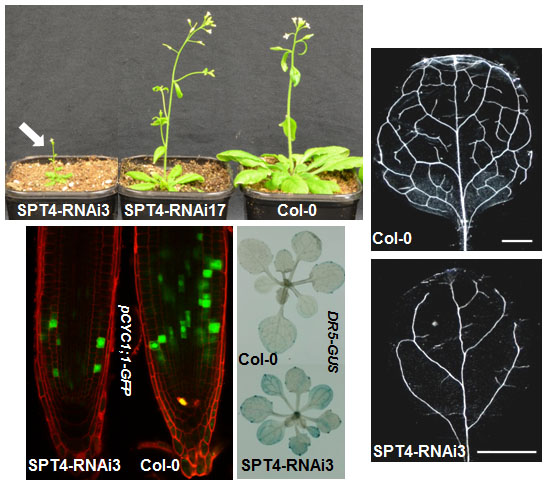
The SPT4 and SPT5 proteins interact in Arabidopsis cells forming the TEF termed SPT4/SPT5 that associates with RNAPII. SPT5 co-localises with elongating RNAPII in the euchromatin and is found in the transcribed regions of actively transcribed genes, but not of inactive genes. Down-regulation of SPT4 expression causes various vegetative and reproductive defects including severely impaired growth resulting from decreased cell proliferation. SPT4-RNAi plants show various auxin-signalling phenotypes (e.g. lateral root density, leaf venation) and a number of auxin-related genes are down-regulated. Consistent with the decreased expression of AUX/IAA genes the SPT4-RNAi plants show an enhanced auxin-response (Dürr et al., 2014 (externer Link, öffnet neues Fenster)). The perturbed development most likely is caused by transcript elongation defects in the SPT4-RNAi plants, as evident from RNAPII accumulation over the transcribed regions of genes.
These examples illustrate the important role of the regulation of transcript elongation in plant developmental processes that we intend to study in more detail in future research.
